Why should people join the Burgess research group?
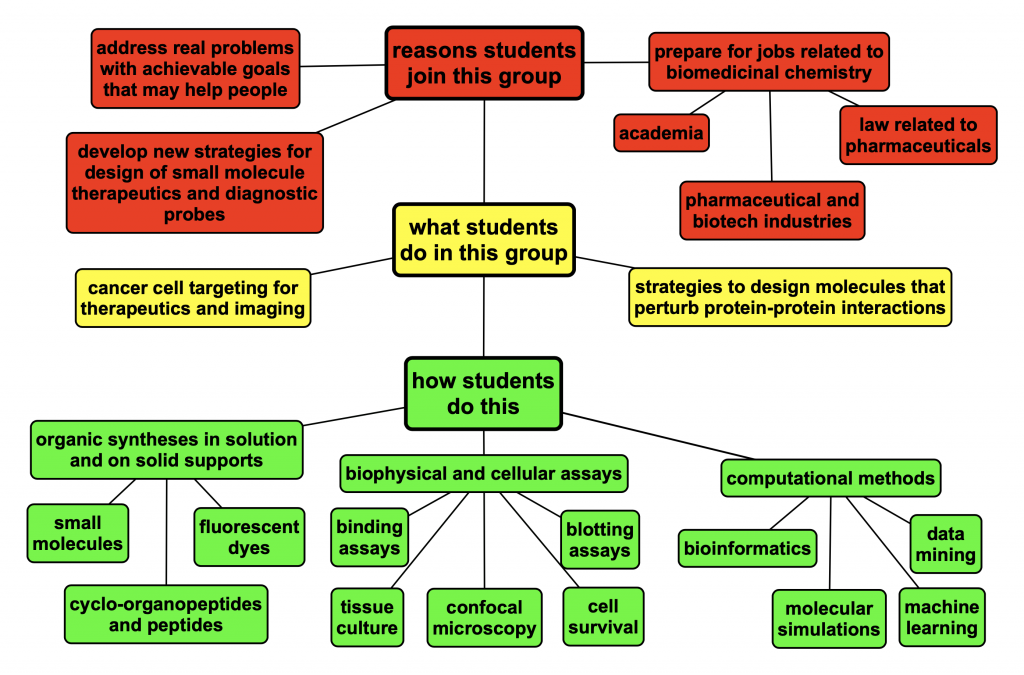
The Burgess Group is for PhD students and postdocs who are interested in applying organic techniques to problems in biomedicinal chemistry involving small molecules that interact with cell surface receptors to impact disease states. We are particularly interested in new methods and approaches that could be used for drug design
what we do
Design new molecules to interact with cell surface proteins on diseased cells so that we can stimulate intracellular signaling, deliver bioactive substances with increased therapeutic indices, or image diseased cells via fluorescence or positron emission tomography.
how we do this
Everybody in our group does molecular design and organic syntheses, though the amount of synthesis can vary from peptides and peptidomimetics only, to more sophisticated molecules. Group members then accumulate combinations of specialist skills that include: biophysical assays (eg fluorescence polarization and ELISA), tissue culture and cellular assays (eg FACS and Western blotting, intracellular imaging), or bioinformatics including data mining and perhaps some machine learning. Many students take courses outside the department of chemistry. Interested students can learn many of these techniques. Prior experience is not required: we can teach you. We collaborate with other research groups for in vivo work.
illustrative current projects
Design of cyclo-organopeptides that interact with Trk receptors to impact dry-eye disease and neurotrophic keratitis.
PROTACs to simultaneously degrade CDKs 2, 4, 6, and indirectly cyclins A, D, and E, to induce apoptosis in cancer cells by suppressing phosphoralation of retinoblastoma protein (Rb).
Design of water-soluble near IR2 dyes for relatively deep tissue imaging of cancer via fluorescence and photoacoustic spectroscopies.
Massively parallel molecular dynamics to match preferred conformations of cyclo-organopeptides with hot spots in protein ligands that are involved in protein-protein interactions.
Development of machine learning methods to identify potential binding pockets on protein surfaces and suggest small molecules that may bind them.
Synthetic Schellman turn and mimics to induce peptide helicity via capping for drug designs involving helical peptides at interfaces.
Stapled helices formed from BODIPY-containing cyclic peptides, and their use in rapid screening of binding to cell surface receptors.